Abstract
We describe the neurological, electrophysiological, and genetic features of autosomal dominant distal hereditary motor neuronopathy (HMN) in a three-generation Dutch family, including 12 patients with distal muscle weakness and atrophy. The severity of disease ranged from disabling muscle weakness to a subclinical phenotype. Neurologic exams of nine patients and nerve conduction studies (NCS) and myography in five endorsed the variable presentations of HMN in this family, including patients with only lower (four), upper (one), or both upper and lower extremities involvement (four). Asymmetrical or strictly unilateral disease was noted in three patients. Three also showed pyramidal features. A genome-wide search combining SNP arrays (250K) with parametric linkage analysis identified a novel locus on chromosome 16p (mLOD = 3.28) spanning 6 Mb (rs6500882–rs7192086). Direct sequencing excluded mutations in the SIMPLE/LITAF gene (mapping to the 16p locus) and identified a pathogenic mutation (p.N88S) in BCLS2 (11q12–q14). All 12 affected relatives had the BSCL2 mutation and the chromosome 16p haplotype and showed features of motor neuron degeneration. One patient had a very mild phenotype with bilateral pes cavus, normal concentric needle electromyography but signs of motor neuron involvement at electrophysiological muscle scan (EMS). Similar EMS abnormalities in addition to abnormal NCS and myography were observed in a clinically unaffected person (carrying only the 16p haplotype). These results expand the clinical spectrum of HMN and suggest a digenic inheritance of HMN in this family with a BSCL2 mutation and a chromosome 16 locus likely contributing to the phenotype.
Similar content being viewed by others
Introduction
Distal hereditary motor neuronopathy (HMN) is defined by progressive muscle wasting and weakness without clinical, electrophysiological, or morphological sensory involvement due to anterior horn cell degeneration and is also referred to as the spinal subtype of hereditary motor and sensory neuropathy (HMSN) or Charcot–Marie–Tooth disease (CMT) [1–3]. According to clinical findings, age at onset, and mode of inheritance, seven subtypes of HMN have been defined (HMN I–VII) [2, 4]. A genetic classification of HMN is gradually evolving with the identification of eight genes and 14 loci, associated with distal HMN (Table 1) [2, 3, 5]. Several of these genes are also associated with HMSN subtypes, hereditary motor neuron diseases, and hereditary spastic paraplegias, indicating an overlapping clinical spectrum. For instance, mutations within the N-glycosylation site of BSCL2 (Bernardinelli–Seip congenital lipodystrophy) gene are believed to cause a gain of function with accumulation of unfolded protein in the endoplasmatic reticulum leading to aggregate formation and neurodegeneration. These mutations are associated with HMN as well as Silver syndrome, an autosomal dominant spastic paraparesis with marked atrophy of the hand muscles [6, 7]. The senataxin gene (SETX), primary associated with autosomal dominant juvenile amyotrophic lateral sclerosis (ALS4) and spastic paraplegia without neuropathy (SPG19), has now also been associated with distal HMN with pyramidal features [3, 8]. Nonsense mutations in senataxin cause ataxia-oculomotor apraxia type 2 (AOA2), a neurodegenerative disorder with cerebellar ataxia and severe axonal neuropathy [9]. Furthermore, mutations in genes encoding small heath shock protein (HSP1, HSP8) were reported in HMN type 2 as well as in two axonal HMSN subtypes, CMT2F and CMT2L. HMN type V and CMT2D, both predominantly involving the hands, appeared to be allelic disorders, with mutations in the glycil tRNA synthetase (GARS) gene [2, 4].
The pathogenic mechanism involved with these different genetic forms of HMN are not well understood and suggest disruption of a variety of cellular processes, including protein misfolding, RNA/DNA transcription, axonal transport, and endoplasmatic reticulum stress.
We present the neurological and electrophysiological characteristics of autosomal dominant HMN in a large Dutch family, displaying variability in localization (with a hand type and a foot type) and co-occurrence of pyramidal signs as well as a wide range in severity of distal muscle weakness. The influence of genetic and/or environmental modifiers has been suggested previously to explain the clinical variability of HMN within families or carriers of a specific mutation [4, 10, 11]. We combined the high-throughput SNP array (250K) technology with parametric linkage analysis and direct sequencing and identified a novel HMN locus on chromosome 16p segregating with a mutation in the BSCL2 gene previously associated with HMN/Silver syndrome. Our results suggest that the novel locus on chromosome 16 may contain a gene modifying BSCL2-associated disease.
Patients and methods
Patients
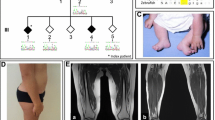
Fifteen members of a Dutch three-generation family were evaluated clinically by one neurologist (EB). The pedigree is depicted in Fig. 1. Nine family members (four men, five women) were considered to be definitely affected. Two individuals, who were reported to be affected (II:3, III:8) could not be examined; however, we were able to obtain medical records confirming the diagnosis. DNA samples of these two patients as well as a sample from stored DNA of a deceased relative who was also reported to be affected (II:1) were obtained. Three asymptomatic relatives did not have HMN features upon neurologic examination. In three clinically affected (II:5, II:7, and III:1) and one clinically unaffected relative (II:9), nerve conduction studies (NCS) and concentric needle myography were performed, including an electrophysiological muscle scan (EMS) in II:5 and II:9. An EMS is a detailed stimulus intensity—compound muscle action potential (CMAP) amplitude response curve—representing the contribution of individual motor units (MUs) to the CMAP. This test was recently developed in our clinic and can detect large MUs due to chronic reinnervation, which might be missed in concentric needle examination [12]. Medical records of two additional patients (III:4 and IV:1) provided results of NCS studies.
Pedigree of the family with distal HMN. Filled symbols indicate affected individuals. A question mark identifies individuals with “undetermined” phenotype. Below each individual, numbered alleles correspond to each of the microsatellite markers: D11S905 (57.4 cM), D11S4191 (64.9 cM), D11S987, (72.2 cM), and D11S1314 (78.75 cM). Marker order and positions are according to the Marshfield sex-averaged genetic map. A black bar indicates the shared haplotype. BSCL2 is located between D11S4191 and D11S987. A positive (plus signs) or negative (minus signs) sign indicate presence or absence of p.N88S BSCL2 mutation. The most likely (non-recombinant) haplotype for III:3 would be (5–3–5–3), indicating no linkage to the BSCL2 region. However, she was also carrying the p.N88S mutation. Several single and double recombinations are displayed
Genotyping
Genomic DNA was isolated from peripheral blood using the Puregene DNA kit (Gentra Systems) [13]. The genome-wide search was conducted in 15 family members (11 affected) using the Affymetrix GeneChip Mapping 250K Nsp Array containing 262,264 SNP markers. Samples were processed according to the manufacturer’s instructions (Affymetrix GeneChip Mapping Assay). Affymetrix GCOS software v1.4, GTYPE software v4.1, and the dynamic model algorithm were used to derive SNP genotypes. Finally, the BRLMM algorithm was used to improve overall performance.
Linkage analysis and locus identification
The statistical package EasyLinkage Plus v5.02 designed to perform automated linkage analyses using large-scale SNP data was used to perform all analyses [13]. PedCheck (v1.1) and Merlin (v1.0) were used to search for unusual patterns of Mendelian inheritance consistent with potential genotyping errors or unusual double recombinants. All SNPs showing inconsistency in transmission were removed from further analyses. Allegro v1.2c software (incorporated in the EasyLinkage Plus v5.02 package) was used to perform fully automated single point and multipoint linkage analysis. LOD scores were obtained using a dominant model of inheritance, with a penetrance of 98%, a phenocopy rate of 1:1,000, and a disease allele frequency of 1:10,000. Allele frequencies of all genotyped SNPs (derived from Caucasian control individuals), map order, and genetic inter-SNPs distances were taken from the Affymetrix website (Marshfield sex-averaged genetic map).
An “affected only” linkage analysis was performed: Individuals with a definite clinical diagnosis were included in the analysis as “affected”. All relatives with insufficient clinical data or no clinical evidence of disease were classified as “diagnosis unknown”, regardless of their age.
Whole genome linkage analysis was performed with predefined spacing of 0.2 to 0.4 cM. Then, single chromosomes showing positive linkage signals were independently analyzed under the same conditions and haplotypes were constructed. Graphical visualization of haplotypes was performed with HaploPainter v029.5 [14].
Microsatellite markers mapping to the identified “candidate” genomic regions were selected. PCRs were performed with M-13 tailed primers and addition of fluorescent labels (FAM, VIC, or NED). Products were run on an ABI Prism 3100 genetic sequencer (Applied Biosystems) and analyzed using the GeneMapper software v.3.0 (Applied Biosystems).
Sequencing analysis
Direct sequencing of the entire coding region and the exon–intron boundaries of candidate genes was undertaken using PCR primers designed by Primer3 software [15]. Amplified PCR products were purified and sequenced using BigDye Terminator chemistry v3.1 on an ABI Prism 3100 genetic analyzer (Applied Biosystems). Sequences were aligned and compared with consensus sequences obtained from the human genome databases, using SeqScape v2.5 [16].
Quantitative PCR
Quantitative PCR was performed on genomic DNA and cDNA from several patients and controls. For each experiment, two housekeeping genes were used as reference genes, and two different assays were performed covering exons 2 to 4 of the LITAF/SIMPLE gene. All samples were done in triplicate, and experiments were repeated at least twice. Reactions were performed using the iTaq Sybr Green supermix with ROX (Bio-Rad) in a 7300 real-time PCR machine from Applied Biosystems.
Results
Clinical findings
The results of the neurologic exams and electrophysiological studies are summarized in Tables 2 and 3. The proband (III:1), evaluated at the age of 49 years, reported slowly progressive weakness and stiffness of the legs from the age of 10 years: At age 30, she needed orthopedic shoes, and at age 48 years, she was walking with a cane. Her deceased mother (II:1) was diagnosed previously with early onset “neurogenic muscle atrophy,” predominantly affecting the upper limbs. Neurological examination of III:1 displayed a combination of spastic and steppage gait. There was a moderate asymmetrical paresis of the distal legs, most prominent in the ankle dorsiflexors with bilateral pes cavus. Hypertonia and hyperreflexia of the lower limbs was found, with bilateral Babinski’s sign. There was no sensory involvement, and examination of the upper extremities was normal. NCS showed markedly reduced CMAP amplitudes in the lower extremities, with some slowing of conduction velocity. Sensory NCS and concentric needle electromyography (EMG) were normal (Table 3). MRI imaging of the brain revealed no abnormalities.
A maternal uncle of the proband (II:7) had never been able to walk on heels or toes. From the age of 54, he had noticed a left foot drop. Examination at age 69 revealed asymmetrical weakness of the ankle dorsiflexors, with bilateral pes cavus, but without pyramidal features. NCS showed reduced to absent CMAP amplitudes in the lower extremities (Table 3). His son (III:8) was reported with weakness and wasting of the hand musculature from the age of 6 years.
Upon the neurological examination of three reported asymptomatic maternal aunts (II:5, II:9, and II:10), in II:5 (aged 73 years), we found bilateral pes cavus and mild weakness of the ankle dorsiflexors without pyramidal signs (Fig. 3). She confirmed that from her teens, she had never been able to wear boots due to her foot deformity, but she had not experienced functional restrictions. Motor NCS and myography were normal. Sensory NCS of the right distal median nerve was indicative of a sensory carpal tunnel syndrome (Table 3). The EMS, recorded at the abductor pollicis brevis muscle, showed several steps up to 500 µV, indicating the presence of large MUs, probably in combination with a decreased number of functional MUs. This is representing a chronic denervation–reinnervation process. Her sisters II:9 and II:10 showed no neurological abnormalities at age 70. In II:9, the distal CMAP amplitude of the right median nerve was reduced, and myography showed a markedly reduced maximal recruitment pattern in the abductor pollicis brevis muscle with giant MU potentials up to 6–7 mV (Table 3). The EMS showed a similar pattern as in her clinically affected sister II:5.
Neurological examination of the asymptomatic sister III:2 of the proband (at age 49) was normal. A younger sister (III:3) of the proband, examined at age 49 years, had noticed a non-disabling weakness of the right hand since the age of 16 years and a mild unilateral foot drop beginning 2 years ago. Neurological examination displayed weakness of intrinsic muscles of the right hand, with atrophy and clawing of the fingers. There was mild weakness of the right ankle dorsiflexors and hyperreflexia of the right leg with a Babinski’s sign.
Onset of hand dysfunction in the youngest sister (III:4) of the proband was at the age of 12 years, and she had noticed a mild gait disturbance beginning several years ago. Neurological examination at age 43 revealed weakness and atrophy of the hand muscles, most prominent in the thenar and first dorsal interosseus muscles (with sparing of the hypothenar muscles: “split-hand” type atrophy), with clawing of the fingers. There was mild weakness of ankle dorsiflexors, without atrophy or pes cavus. Hyperreflexia of the legs with bilateral Babinski’s signs were present. Previous electrophysiological studies at age 24 showed mild slowing of motor nerve conduction in the arms (Table 3).
A maternal uncle of the proband (II:3) and his son (III:6) both reported a non-disabling childhood onset bilateral pes cavus. Neurological examination of III:6 at the age of 46 years revealed distal weakness and atrophy of the legs, most pronounced at the ankle dorsiflexors with a steppage gait without pyramidal features.
Both IV:1 and IV:2, examined at the age of 16 and 12 years, respectively, displayed a split-hand type atrophy of the hand musculature, similar to the symptoms of their mother (III:4). These two patients lacked pyramidal signs. IV:1 also displayed mild weakness of the ankle dorsiflexors. NCS in IV:1 at age 14 showed reduced to absent CMAP amplitudes in upper and lower extremities. Myography of tibialis and first interosseus dorsalis muscles showed giant MU potentials (Table 3). Medical history of IV:1 reported seizures from the age of 2 to 7 years. IV:2 was diagnosed with a disorder in the attention-deficit hyperactivity spectrum.
Onset of symptoms in IV:3 was at the age of 11 years. Neurological examination at age 16 revealed mild weakness and atrophy of the intrinsic hand muscles and severe weakness of the opponens pollicis muscles and the ankle dorsiflexors, with bilateral pes cavus, without pyramidal features.
Analysis of genes previously associated with HMN
Direct sequencing of the coding region of the GARS, HSP1, HSPB8 and Neurofilament Protein, Light polypeotide (NEFL) genes excluded mutations in the index patient (Neurogenetics Laboratory, Amsterdam). The Senataxin (SETX) locus on 9q34 was excluded by haplotype analyses using eight affected relatives (data not shown). Transmission of microsatellite markers around the Seipin (BSCL2) locus on 11q13 indicated that all but one of the 12 patients in this family shared the same haplotype, making linkage to this locus unlikely, since this would require the occurrence of at least one double recombinant and several single recombinations in one patient and her relatives (Fig. 1). Although neither the haplotype nor the linkage analysis (LOD score of 2.1, Table 4) supported a causative role of BSCL2, we decided to pursue investigations of this gene. The phenotype of this family was similar to previously described Dutch families with Silver syndrome [6]. Sequencing of exon 3 of the BSCL2 gene detected a known heterozygous c.263A > G (p.N88S) mutation [7] present in all 12 patients but not found in the three asymptomatic relatives II:10, III:2, and in II:9 (with abnormal electrophysiological and EMS studies).
Linkage analysis and haplotyping
We performed a genome-wide linkage analysis in 15 relatives, including 11 patients, using the Affymetrix 250 K Nsp arrays. After setting inter-marker spacing to 0.2, 0.3, and 0.4 cM, a total of 15,473, 10,622, and 8,090 SNPs were selected by the program for further linkage analyses, respectively. When multipoint linkage analyses were performed, several genomic regions displayed LOD scores above 2 (Table 4). The highest LOD scores (2.7) were observed on chromosome 16 and 17. We tested microsatellite markers covering these regions (D16S404, D16S519, D16S3075, D16S680, D16S3103 and D17S1857, D17S798, D17S1880, D17S1850, D17S1868). Haplotype analysis revealed that all 12 patients were sharing the same “disease” haplotype on chromosome 16p (Fig. 2), indicating complete segregation of HMN with the chromosome 16 locus. Based on the SNP data, the haplotype on 16p13.3–p13.12 spans approximately 14.6 cM (6 Mb) between the SNPs rs6500882 (6.9 Mb, SNP_A2078682) and rs7192086 (12.96 Mb, SNP_A1813744). Multipoint linkage analyses using the microsatellite data yielded a significant mLOD of 3.28 for chromosome 16p (using the same genetic model described before).
Pedigree of family with HMN showing SNP haplotypes corresponding to the 16p13.3–p13.12 region. Filled symbols indicate affected individuals. A question mark identifies individuals with undetermined phenotype used in the analysis. Informative SNPs with a spacing of 0.3 cM has been used in this figure. Haplotypes corresponding to individuals I:1, I:2, II:2, and II:8 were reconstructed by the program (Allegro). A dark brown bar identifies the disease haplotype. Recombinations occurring in III:8 and III:6 are delimiting the area of 14.6 cM to approximately 12 cM (6 Mb) between SNP_A2078682 or rs6500882 (not depicted in figure due to the selected spacing) and SNP_A1813744 or rs7192086
The identified chromosome 16p locus (from 6.9 Mb until 12.96 Mb) contains 49 genes. One of them, the LITAF/SIMPLE gene, associated with CMT1C, was an obvious candidate for further analysis. Direct sequencing of the four exons plus part of the 5′ and 3′ UTR of the gene displayed only two DNA variants in the coding region of the gene Ile92Val (rs4280262), a known polymorphism and a novel silent change (p.Asn110Asn). Both variants were not co-segregating with the phenotype in this family and therefore are likely not pathogenic.
We then investigated whether copy number variants within LITAF/SIMPLE could have a role in causing the HMN phenotype. We performed quantitative real-time PCR on genomic DNA from four patients (III:1, III:3, III:4, and IV:1) and compared them to unaffected family members and controls. No evidence for deletions or duplication was found along the coding region of LITAF. Quantitative real-time PCR performed on cDNA detected upregulation (3-fold increase) in LITAF levels in two patients (III:1 and III:4) and in one unaffected (III:2) family member, while two other patients (III:6 and IV:3) and two asymptomatic relatives (II:9 and II:10) had normal levels. These results showed that changes in LITAF expression do not influence disease in this family.
Discussion
We described an extensive family with autosomal dominant distal HMN showing a wide range in the severity and localization of neuromuscular symptoms. In nine examined patients, distribution of muscle weakness and atrophy included involvement of predominantly the feet (n = 4), the hands (n = 1), or both upper and lower extremities (n = 4), often with split-hand type atrophy. Co-occurrence of pyramidal features was observed in three patients. In two patients, muscle involvement was asymmetric, and in one, strictly unilateral muscle weakness and atrophy was observed. Severity of the disease ranged from adolescent patients with disabling muscle weakness to an elderly patient with only mild weakness of the ankle dorsiflexors and bilateral pes cavus (II:5) (Fig. 3). The asymmetrical involvement of muscle weakness, the pyramidal signs, and the absence of sensory deficits indicate spinal anterior horn cells involvement, as was supported by electrophysiology showing signs of chronic motor axonal injury with normal sensory conduction.
The recently introduced and promising technique of the EMS [12] showed signs of chronic motor neuron involvement in the upper extremities in one patient (II:5) with a relatively mild phenotype, whereas motor NCS studies and myography were normal in this patient. The EMS may therefore be considered as a sensitive technique to detect subtle or even subclinical motor neuron dysfunction. This should be confirmed with further NCS and EMS studies in affected and healthy relatives.
In this family, one of the two reported mutations in exon 3 of the BSCL2/Seipin gene associated with HMN and Silver syndrome was identified in all 12 clinically affected individuals. Functional (in vitro) evidence is indicating that the N88S and S90L mutations destroy a predicted N-glycosylation site of the protein at positions 88–90, resulting in aggregates formation, cell death, and neurodegeneration [7, 17]
Van de Warrenburg et al. describe two other Dutch families with the same c.236A > G mutation, showing a similar age at onset and intrafamilial variability in localization as observed in our family [6]. However, the unilateral involvement observed in this study has not been described previously. As illustrated in Tables 2 and 3, in the majority of the patients, onset of clinical features occurs at age 10–16 years. However, the three patients of the youngest generation seem to display a severe phenotype considering their age, mostly with bilateral involvement of both hand and feet. This suggestion of possible anticipation could not be explained by the 16p haplotype shared by all affected relatives.
Auer-Grumbach et al. described the variability of the clinical phenotype in 90 patients with the p.N88S mutation, indicating that approximately 25% of the patients were not affected or were subclinically affected, sometimes only displaying NCS abnormalities. In addition, patients with this specific mutation have been diagnosed as a hand-type d-HMN (31.1%), classical Silver syndrome (14.5%), CMT-phenotype (20%), or pure spastic paraparesis (10%) [18].
This extreme variability in phenotype in a disorder related so far to only two mutations in the BSCL2 gene is remarkable and suggests influence of genetic or environmental modifiers. The digenic inheritance of the pathogenic mutation in the BSCL2 gene and the locus on chromosome 16p suggests that the latter may contain a gene-modifying disease expression in this family. The 16p region was shared by 14 relatives: 12 definite patients and two clinically unaffected twin sisters II:9 and II:10 without the p.N88S mutation. Electrophysiological studies in the unaffected II:9 did show motor neuron damage with a diminished CMAP amplitude of the median nerve and signs of reinnervation in myography. These abnormalities are not likely to be attributable to an asymptomatic carpal tunnel syndrome because sensory NCS of the median nerve were normal. Furthermore, the EMS in II:9 showed similar signs of motor neuron involvement as in her mildly affected sister II:5, who is carrying both the BLCS2 mutation and the chromosome 16p haplotype. These findings suggest that the 16p locus may be associated with attenuated forms of HMN. Alternatively, these observations may indicate that the 16p locus is not sufficient for disease causing but has additive pathogenic effect.
An obvious candidate gene, the LITAF/SIMPLE gene, maps to the 16p region. The gene was associated with a demyelinating HMSN-subtype (HMSN1C). Also, a CMT2 phenotype, displaying asymmetric distal muscle weakness and normal NCV, has been associated with a LITAF mutation [19]. In our family, mutations in the coding region of the LITAF/SIMPLE gene were excluded by direct sequencing, and no copy number variants (genomic DNA) were detected. Differences in gene expression (RNA-cDNA) were found but were not related to disease status.
The Ingenuity database (Ingenuity Systems, www.ingenuity.com) identified 33 of the 49 genes located on the chromosome 16 locus. Among those 33 genes, there are some possible candidate genes involved in DNA damage response and double-strand break repair pathways previously associated with neurodegeneration (i.e., CARHSP1 (calcium-regulated heat-stable protein), GRIN2A (N-methyl- d -aspartate glutamate receptor 2A), SOCS1 (suppressor of cytokine signaling 1), SNN (stannin), and TNFRSF17 (tumor necrosis factor receptor superfamily, member 17), A2BP1 (ataxin 2-binding protein 1), CIITA (class II, major histocompatibility complex, transactivator), and RSL1D1 (ribosomal L1 domain containing 1)).
In conclusion, clinical and molecular studies of a large Dutch family with HMN detected a concomitant pathogenic mutation p.N88S in the BSCL2 gene and linkage to a locus on chromosome 16p associated with a wide variation in severity and presentation of the neuromuscular features. The role of the locus on chromosome 16p in HMN pathogenesis remains to be elucidated; it may act as a disease modifier, possibly explaining the phenotypic variability ranging from strictly neuropathic weakness to a spastic paraplegia.
References
Auer-Grumbach M, Löscher WN, Wagner K, Petek E, Körner E, Offenbacher H, Hartung HP (2000) Phenotypic and genotypic heterogeneity in hereditary motor neuronopathy type V: a clinical, electrophysiological and genetic study. Brain 123(Pt 8):1612–1623. doi:10.1093/brain/123.8.1612
Harding AE (1993) Inherited neuronal atrophy and degeneration predominantly of lower motor neurons. In: Dyck TP, Griffin JW, Low PA, Poduslo JF (eds) Peripheral neuropathy. Saunders, Philadelphia, pp 1051–1064
Spinal Muscular Atrophy database at http://www.neuro.wustl.edu/neuromuscular/synmot.html#distalsma. Accessed May 2008
Irobi J, De Jonghe P, Timmerman V (2004) Molecular genetics of distal hereditary motor neuropathies. Hum Mol Genet 13(2):R195–R202
Maystadt I, Rezsöhazy R, Barkats M, Duque S, Vannuffel P, Remacle S, Lambert B, Najimi M, Sokal E, Munnich A, Viollet L, Verellen-Dumoulin C (2007) The nuclear factor kappaB-activator gene PLEKHG5 is mutated in a form of autosomal recessive lower motor neuron disease with childhood onset. Am J Hum Genet 81:67–76. doi:10.1086/518900
van de Warrenburg BP, Scheffer H, van Eijk JJ, Versteeg MH, Kremer H, Zwarts MJ, Schelhaas HJ, Van Engelen BG (2006) BSCL2 mutations in two Dutch families with overlapping Silver syndrome-distal hereditary motor neuropathy. Neuromuscul Disord 16:122–125. doi:10.1016/j.nmd.2005.11.003
Windpassinger C, Auer-Grumbach M, Irobi J, Patel H, Petek E, Hörl G, Malli R, Reed JH, Dierick I, Verpoorten N, Warner TT, Proukakis C, Van Den Bergh P, Verellen C, Van Maldergem L, Merlini L, De Jonghe P, Timmerman V, Crosby AH, Wagner K (2004) Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat Genet 36:271–276. doi:10.1038/ng1313
De Jonghe P, Auer-Grumbach M, Irobi J, Wagner K, Plecko B, Kennerson M, Zhu D, De Vriendt E, Van Gerwen V, Nicholson G, Hartung HP, Timmerman V (2002) Autosomal dominant juvenile amyotrophic lateral sclerosis and distal hereditary motor neuronopathy with pyramidal tract signs: synonyms for the same disorder? Brain 125:1320–1325. doi:10.1093/brain/awf127
Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, Nakamura J, Gatei M, Criscuolo C, Filla A, Chessa L, Fusser M, Epe B, Gueven N, Lavin MF (2007) Senataxin, defective in ataxia oculomotor apraxia type2, is involved in the defense against oxidative DNA damage. J Cell Biol 177:969–979. doi:10.1083/jcb.200701042
Cafforio G, Calabrese R, Morelli N, Mancusco M, Piazza S, Martinuzzi A, Bassi MT, Crippa F, Siciliano G (2008) The first Italian family with evidence of pyramidal impairment as phenotypic manifestation of Silver syndrome BSCL2 gene mutation. Neurol Sci 29:189–191. doi:10.1007/s10072-008-0937-y
Irobi J, Van den Bergh P, Merlini L, Verellen C, van Maldergem L, Dierick I, Verpoorten N, Jordanova A, Windpassinger C, De Vriendt E, Van Gerwen V, Auer-Grumbach M, Wagner K, Timmerman V, De Jonghe P (2004) The phenotype of motor neuropathies associated with BSCL2 mutations is broader than Silver syndrome and distal HMN type V. Brain 127:2124–2130. doi:10.1093/brain/awh232
Blok JH, Ruitenberg A, Maathuis EM, Visser GH (2007) The electrophysiological muscle scan. Muscle Nerve 36:436–446. doi:10.1002/mus.20838
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. doi:10.1093/nar/16.3.1215
Hoffmann K, Lindner TH (2005) easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics 21:3565–3567. doi:10.1093/bioinformatics/bti571
Primer3 software. http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi
UCSC Genome Bioinformatics. http://genome.ucsc.edu, http://www.ncbi.nlm.nih.gov
Ito D, Suzuki N (2007) Molecular pathogenesis of seipin/BSCL2-related motor neuron diseases. Ann Neurol 61:237–250. doi:10.1002/ana.21070
Auer-Grumbach M, Schlotter-Weigel B, Lochmüller H, Strobl-Wildemann G, Auer-Grumbach P, Fischer R, Offenbacher H, Zwick EB, Robl T, Hartl G, Hartung HP, Wagner K, Windpassinger C (2005) Phenotypes of the N88S Bernardinelli-Seip congenital lipodystrophy 2 mutation. Ann Neurol 57:415–424. doi:10.1002/ana.20410
Saifi GM, Szigeti K, Wiszniewski W, Shy ME, Krajewski K, Hausmanowa-Petrusewicz I, Kochanski A, Reeser S, Mancias P, Butler I, Lupski JR (2005) SIMPLE mutations in Charcot-Marie-Tooth disease and the potential role of its protein product in protein degradation. Hum Mutat 25:372–383. doi:10.1002/humu.20153
Acknowledgments
We thank all family members for their participation in the study. We are grateful to Dr. Frank Baas (Neurogenetics lab, Academic Medical Centrum, Amsterdam University) for performing molecular diagnosis of candidate genes in the index patient. We acknowledge Tom de Vries Lentsch for the artwork. Support was given by a grant from the Center for Biomedical Genetics.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Brusse, E., Majoor-Krakauer, D., de Graaf, B.M. et al. A novel 16p locus associated with BSCL2 hereditary motor neuronopathy: a genetic modifier?. Neurogenetics 10, 289–297 (2009). https://doi.org/10.1007/s10048-009-0193-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-009-0193-1