Abstract
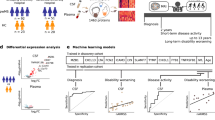
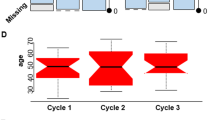
We employ shotgun proteomics and data-independent acquisition (DIA) mass spectrometry to analyze cerebrospinal fluid longitudinally collected from 14 amyotrophic lateral sclerosis (ALS) patients (8 males and 6 females). We perform three main analyses of these data: (1) examine the intra- and inter-patient protein variability in CSF; (2) explore the association of inflammation with rate of disease progression; and (3) develop a mixed-effects model to best explain the decrease in ALS-Functional Rating Scale (ALS-FRS) score. Overall, the CSF protein abundances are tightly regulated with the intra-individual variability contributing just 4% to the overall variance. In four patients, a moderately significant correlation (p < 0.1) was observed between inflammation and rate of disease progression. Using a least absolute shrinkage and selection operator (LASSO) variable selection, we selected 55 viable peptides for mathematical modeling via a linear mixed-effects regression. We then employed forward selection to generate a final model by minimizing Akaike’s information criterion (AIC). The final model utilized changes in abundance from 28 peptides as fixed effects to model progression of the disease in these patients. These peptides were from proteins involved in stress response and innate immunity.

Graphical abstract






Similar content being viewed by others
Abbreviations
- ABC:
-
Ammonium bicarbonate
- AIC:
-
Akaike’s information criterion
- ALS:
-
Amyotrophic lateral sclerosis
- ALS-FRS:
-
ALS-Functional Rating Scale
- CHI3L1:
-
Chitinase-3-like protein 1
- CHI3L2:
-
Chitinase-3-like protein 2
- CHIT1:
-
Chitotriosidase
- CSF:
-
Cerebrospinal fluid
- DDA:
-
Data-dependent acquisition
- DIA:
-
Data-independent acquisition
- DTT:
-
Dithiothreitol
- IAM:
-
Iodoacetamide
- LASSO:
-
Least absolute shrinkage and selection operator
- SDC:
-
Sodium deoxycholate
References
Zufiría M, Gil-Bea FJ, Fernández-Torrón R, Poza JJ, Muñoz-Blanco JL, Rojas-García R, et al. ALS: a bucket of genes, environment, metabolism and unknown ingredients. Prog Neurobiol. 2016;142:104–29.
Boillée S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59.
Gagliardi D, Meneri M, Saccomanno D, Bresolin N, Comi GP, Corti S. Diagnostic and prognostic role of blood and cerebrospinal fluid and blood neurofilaments in amyotrophic lateral sclerosis: a review of the literature. Int J Mol Sci. 2019;20:4152.
Petrov D, Mansfield C, Moussy A, Hermine O. ALS Clinical Trials Review: 20 years of failure. Are we any closer to registering a new treatment? Front Aging Neurosci. 2017;9:68.
Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. Longo DL, editor. N Engl J Med. 2017;377:162–72.
Zarei S, Carr K, Reiley L, Diaz K, Guerra O, Altamirano PF, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int. 2015;6.
ALS Functional Rating Scale. Available from: https://www.outcomes-umassmed.org/ALS/alsscale.aspx. Accessed 7 Feb 2020.
Cedarbaum JM, Stambler N, Malta E, Fuller C, Hilt D, Thurmond B, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci. 1999;169:13–21.
Turner MR, Kiernan MC, Leigh PN, Talbot K. Biomarkers in amyotrophic lateral sclerosis. Lancet Neurol. 2009;8:94–109.
Bereman MS, Beri J, Enders JR, Nash T. Machine learning reveals protein signatures in CSF and plasma fluids of clinical value for ALS. Sci Rep. 2018;8:1–14.
Lind A-L, Wu D, Freyhult E, Bodolea C, Ekegren T, Larsson A, et al. A multiplex protein panel applied to cerebrospinal fluid reveals three new biomarker candidates in ALS but none in neuropathic pain patients. PLoS One. 2016;11:e0149821.
Collins MA, An J, Hood BL, Conrads TP, Bowser RP. Label-free LC-MS/MS proteomic analysis of cerebrospinal fluid identifies protein/pathway alterations and candidate biomarkers for amyotrophic lateral sclerosis. J Proteome Res. 2015;14:4486–501.
Poesen K, Van Damme P. Diagnostic and prognostic performance of neurofilaments in ALS. Front Neurol. 2019;9:1167.
Lu C-H, Petzold A, Topping J, Allen K, Macdonald-Wallis C, Clarke J, et al. Plasma neurofilament heavy chain levels and disease progression in amyotrophic lateral sclerosis: insights from a longitudinal study. J Neurol Neurosurg Psychiatry. 2015;86:565–73.
Benatar M, Wuu J, Lombardi V, Jeromin A, Bowser R, Andersen PM, et al. Neurofilaments in pre-symptomatic ALS and the impact of genotype. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20:538–48.
Varghese AM, Sharma A, Mishra P, Vijayalakshmi K, Harsha HC, Sathyaprabha TN, et al. Chitotriosidase - a putative biomarker for sporadic amyotrophic lateral sclerosis. Clin Proteomics. 2013;10:19.
Thompson AG, Gray E, Thézénas M-L, Charles PD, Evetts S, Hu MT, et al. Cerebrospinal fluid macrophage biomarkers in amyotrophic lateral sclerosis. Ann Neurol. 2018;83:258–68.
Lu C-H, Macdonald-Wallis C, Gray E, Pearce N, Petzold A, Norgren N, et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84:2247–57.
McCombe PA, Pfluger C, Singh P, Lim CYH, Airey C, Henderson RD. Serial measurements of phosphorylated neurofilament-heavy in the serum of subjects with amyotrophic lateral sclerosis. J Neurol Sci. 2015;353:122–9.
Hawkridge AM, Muddiman DC. Mass spectrometry–based biomarker discovery: toward a global proteome index of individuality. Annu Rev Anal Chem (Palo Alto, Calif). 2009;2:265–77.
Enroth S, Johansson Å, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun. 2014;5:1–11.
Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, et al. Strong rules for discarding predictors in lasso-type problems: strong rules for discarding predictors. J R Stat Soc Ser B Stat Methodol. 2012;74:245–66.
Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62.
Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX, et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun. 2018;9:1–12.
Amodei D, Egertson J, MacLean BX, Johnson R, Merrihew GE, Keller A, et al. Improving precursor selectivity in data-independent acquisition using overlapping windows. J Am Soc Mass Spectrom. 2019;30:669–84.
PXD000954 - the pan-human library: a repository of assays to quantify 10 000 proteins by SWATH-MS / SWATH-MS validation data - OmicsDI. Available from: https://www.omicsdi.org/dataset/pride/PXD000954. Accessed 25 April 2019.
Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. 2019.
Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–23.
Wewer Albrechtsen NJ, Geyer PE, Doll S, Treit PV, Bojsen-Møller KN, Martinussen C, et al. Plasma proteome profiling reveals dynamics of inflammatory and lipid homeostasis markers after Roux-en-Y gastric bypass surgery. Cell Syst. 2018;7:601–612.e3.
Gille B, De Schaepdryver M, Goossens J, Dedeene L, De Vocht J, Oldoni E, et al. Serum neurofilament light chain levels as a marker of upper motor neuron degeneration in patients with amyotrophic lateral sclerosis. Neuropathol Appl Neurobiol. 2019;45:291–304.
Canterbury JD, Merrihew GE, MacCoss MJ, Goodlett DR, Shaffer SA. Comparison of data acquisition strategies on quadrupole ion trap instrumentation for shotgun proteomics. J Am Soc Mass Spectrom. 2014;25:2048–59.
Bruderer R, Bernhardt OM, Gandhi T, Xuan Y, Sondermann J, Schmidt M, et al. Optimization of experimental parameters in data-independent mass spectrometry significantly increases depth and reproducibility of results. Mol Cell Proteomics. 2017;16:2296–309.
Lin L, Zheng J, Zheng F, Cai Z, Yu Q. Advancing serum peptidomic profiling by data-independent acquisition for clear-cell renal cell carcinoma detection and biomarker discovery. J Proteome. 2020;215:103671.
Barkovits K, Linden A, Galozzi S, Schilde L, Pacharra S, Mollenhauer B, et al. Characterization of cerebrospinal fluid via data-independent acquisition mass spectrometry. J Proteome Res. 2018;17:3418–30.
Muntel J, Xuan Y, Berger ST, Reiter L, Bachur R, Kentsis A, et al. Advancing urinary protein biomarker discovery by data-independent acquisition on a quadrupole-Orbitrap mass spectrometer. J Proteome Res. 2015;14:4752–62.
Corzett TH, Fodor IK, Choi MW, Walsworth VL, Turteltaub KW, McCutchen-Maloney SL, et al. Statistical analysis of variation in the human plasma proteome. J Biomed Biotechnol. 2010;258494.
Geyer PE, Wewer Albrechtsen NJ, Tyanova S, Grassl N, Iepsen EW, Lundgren J, et al. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol Syst Biol. John Wiley & Sons, Ltd. 2016;12:901.
Harney DJ, Hutchison AT, Hatchwell L, Humphrey SJ, James DE, Hocking S, et al. Proteomic analysis of human plasma during intermittent fasting. J Proteome Res. American Chemical Society. 2019;18:2228–40.
Hühmer AF, Biringer RG, Amato H, Fonteh AN, Harrington MG. Protein analysis in human cerebrospinal fluid: physiological aspects, current progress and future challenges. Dis Markers. 2006;22:3–26.
Reiber H. Proteins in cerebrospinal fluid and blood: barriers, CSF flow rate and source-related dynamics. Restor Neurol Neurosci. IOS Press. 2003;21:79–96.
Reiber H. Dynamics of brain-derived proteins in cerebrospinal fluid. Clin Chim Acta. 2001;310:173–86.
Piehowski PD, Petyuk VA, Orton DJ, Xie F, Moore RJ, Ramirez-Restrepo M, et al. Sources of technical variability in quantitative LC–MS proteomics: human brain tissue sample analysis. J Proteome Res. 2013;12:2128–37.
Stoop MP, Coulier L, Rosenling T, Shi S, Smolinska AM, Buydens L, et al. Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples*. Mol Cell Proteomics. 2010;9:2063–75.
Liu J, Wang F. Role of neuroinflammation in amyotrophic lateral sclerosis: cellular mechanisms and therapeutic implications. Front Immunol. 2017;8:1005.
Komine O, Yamanaka K. Neuroinflammation in motor neuron disease. Nagoya J Med Sci. 2015;77:537–49.
Hooten KG, Beers DR, Zhao W, Appel SH. Protective and toxic neuroinflammation in amyotrophic lateral sclerosis. Neurotherapeutics. 2015;12:364–75.
Shibata, R. An Optimal Selection of Regression Variables. Biometrika. 1981;68:45–54.
Altman N, Krzywinski M. Regression diagnostics. Nat Methods. 2016;13:385–6.
Brettschneider J, Mogel H, Lehmensiek V, Ahlert T, Süssmuth S, Ludolph AC, et al. Proteome analysis of cerebrospinal fluid in amyotrophic lateral sclerosis (ALS). Neurochem Res. 2008;33:2358–63.
Ranganathan S, Williams E, Ganchev P, Gopalakrishnan V, Lacomis D, Urbinelli L, et al. Proteomic profiling of cerebrospinal fluid identifies biomarkers for amyotrophic lateral sclerosis. J Neurochem. 2005;95:1461–71.
Acknowledgments
We gratefully acknowledge the NEALS Biorepository for providing all the biofluids from ALS patients used in this study. We also acknowledge the ALS Association (grant #19-SI-458) for the funding of this project. All mass spectrometry measurements were made in the Molecular Education, Technology, and Research Innovation Center (METRIC) at NC State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The NEALS Biorepository collects and stores cryopreserved CSF from consenting patients, who have been worked up in neurological detail, at the NEALS-affiliated clinical sites. The sample collection and the corresponding clinical information storage and sharing are performed under approved IRB protocols and in accordance with HIPPA.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 624B kb).
Rights and permissions
About this article
Cite this article
Mellinger, A.L., Griffith, E.H. & Bereman, M.S. Peptide variability and signatures associated with disease progression in CSF collected longitudinally from ALS patients. Anal Bioanal Chem 412, 5465–5475 (2020). https://doi.org/10.1007/s00216-020-02765-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02765-8