Abstract
Intronic hexanucleotide expansions in C9ORF72 are common in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia, but it is unknown whether loss of function, toxicity by the expanded RNA or dipeptides from non-ATG-initiated translation are responsible for the pathophysiology. We determined the interactome of C9ORF72 in motor neurons and found that C9ORF72 was present in a complex with cofilin and other actin binding proteins. Phosphorylation of cofilin was enhanced in C9ORF72-depleted motor neurons, in patient-derived lymphoblastoid cells, induced pluripotent stem cell–derived motor neurons and post-mortem brain samples from ALS patients. C9ORF72 modulates the activity of the small GTPases Arf6 and Rac1, resulting in enhanced activity of LIM-kinases 1 and 2 (LIMK1/2). This results in reduced axonal actin dynamics in C9ORF72-depleted motor neurons. Dominant negative Arf6 rescues this defect, suggesting that C9ORF72 acts as a modulator of small GTPases in a pathway that regulates axonal actin dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Renton, A.E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Gijselinck, I. et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 11, 54–65 (2012).
Majounie, E. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330 (2012).
Ciura, S. et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol. 74, 180–187 (2013).
Ling, S.C., Polymenidou, M. & Cleveland, D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Donnelly, C.J. et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013).
Haeusler, A.R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014).
Zu, T. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA 110, E4968–E4977 (2013).
Wen, X. et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014).
Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013).
Therrien, M., Rouleau, G.A., Dion, P.A. & Parker, J.A. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One 8, e83450 (2013).
Jiang, J. et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90, 535–550 (2016).
Koppers, M. et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol. 78, 426–438 (2015).
Burberry, A. et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci. Transl. Med. 8, 347ra93 (2016).
O'Rourke, J.G. et al. C9orf72 is required for proper macrophage and microglial function in mice. Science 351, 1324–1329 (2016).
Lagier-Tourenne, C. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA 110, E4530–E4539 (2013).
Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999).
Sudria-Lopez, E. et al. Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol. 132, 145–147 (2016).
Sullivan, P.M. et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol. Commun. 4, 51 (2016).
Keilhauer, E.C., Hein, M.Y. & Mann, M. Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE-MS) rather than affinity purification mass spectrometry (AP-MS). Mol. Cell. Proteomics 14, 120–135 (2015).
May, S. et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 128, 485–503 (2014).
Flynn, K.C. et al. ADF/cofilin-mediated actin retrograde flow directs neurite formation in the developing brain. Neuron 76, 1091–1107 (2012).
Stern, S. et al. The transcription factor serum response factor stimulates axon regeneration through cytoplasmic localization and cofilin interaction. J. Neurosci. 33, 18836–18848 (2013).
Hornburg, D. et al. Deep proteomic evaluation of primary and cell line motoneuron disease models delineates major differences in neuronal characteristics. Mol. Cell. Proteomics 13, 3410–3420 (2014).
Bravo-Cordero, J.J., Magalhaes, M.A., Eddy, R.J., Hodgson, L. & Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 14, 405–415 (2013).
Moriyama, K., Iida, K. & Yahara, I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1, 73–86 (1996).
Jablonka, S., Beck, M., Lechner, B.D., Mayer, C. & Sendtner, M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J. Cell Biol. 179, 139–149 (2007).
Agnew, B.J., Minamide, L.S. & Bamburg, J.R. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270, 17582–17587 (1995).
Nunoue, K., Ohashi, K., Okano, I. & Mizuno, K. LIMK-1 and LIMK-2, two members of a LIM motif-containing protein kinase family. Oncogene 11, 701–710 (1995).
Yang, N. et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809–812 (1998).
Levine, T.P., Daniels, R.D., Gatta, A.T., Wong, L.H. & Hayes, M.J. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29, 499–503 (2013).
Zhang, D., Iyer, L.M., He, F. & Aravind, L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3, 283 (2012).
Al-Awar, O., Radhakrishna, H., Powell, N.N. & Donaldson, J.G. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol. Cell. Biol. 20, 5998–6007 (2000).
Iden, S. & Collard, J.G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 9, 846–859 (2008).
Hernández-Deviez, D.J., Roth, M.G., Casanova, J.E. & Wilson, J.M. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol. Biol. Cell 15, 111–120 (2004).
Radhakrishna, H., Klausner, R.D. & Donaldson, J.G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 134, 935–947 (1996).
Bourmoum, M., Charles, R. & Claing, A. The GTPase ARF6 controls ROS production to mediate angiotensin II-induced vascular smooth muscle cell proliferation. PLoS One 11, e0148097 (2016).
Cingolani, L.A. et al. Activity-dependent regulation of synaptic AMPA receptor composition and abundance by β3 integrins. Neuron 58, 749–762 (2008).
Huang, W. et al. mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat. Neurosci. 16, 441–448 (2013).
Fratta, P. et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 126, 401–409 (2013).
Bernard, O. Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39, 1071–1076 (2007).
Santy, L.C. & Casanova, J.E. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599–610 (2001).
Lewis-Saravalli, S., Campbell, S. & Claing, A. ARF1 controls Rac1 signaling to regulate migration of MDA-MB-231 invasive breast cancer cells. Cell. Signal. 25, 1813–1819 (2013).
Franssen, E.H. et al. Exclusion of integrins from CNS axons is regulated by Arf6 activation and the AIS. J. Neurosci. 35, 8359–8375 (2015).
Farg, M.A. et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 23, 3579–3595 (2014).
Innocenti, M. et al. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160, 17–23 (2003).
Innocenti, M. et al. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 156, 125–136 (2002).
Wiese, S. et al. Isolation and enrichment of embryonic mouse motoneurons from the lumbar spinal cord of individual mouse embryos. Nat. Protoc. 5, 31–38 (2010).
Rogers, M.L. et al. Functional monoclonal antibodies to p75 neurotrophin receptor raised in knockout mice. J. Neurosci. Methods 158, 109–120 (2006).
Subramanian, N. et al. Role of Nav1.9 in activity-dependent axon growth in motoneurons. Hum. Mol. Genet. 21, 3655–3667 (2012).
Götz, R. et al. Bag1 is essential for differentiation and survival of hematopoietic and neuronal cells. Nat. Neurosci. 8, 1169–1178 (2005).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Michalski, A. et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics 11, O111.013698 (2012).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2014).
Japtok, J. et al. Stepwise acquirement of hallmark neuropathology in FUS-ALS iPSC models depends on mutation type and neuronal aging. Neurobiol. Dis. 82, 420–429 (2015).
Reinhardt, P. et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12, 354–367 (2013).
Lojewski, X. et al. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum. Mol. Genet. 23, 2005–2022 (2014).
Wächter, N., Storch, A. & Hermann, A. Human TDP-43 and FUS selectively affect motor neuron maturation and survival in a murine cell model of ALS by non-cell-autonomous mechanisms. Amyotroph. Lateral Scler. Frontotemporal Degener. 16, 431–441 (2015).
Reinhardt, P. et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One 8, e59252 (2013).
Fischer, M., Kaech, S., Knutti, D. & Matus, A. Rapid actin-based plasticity in dendritic spines. Neuron 20, 847–854 (1998).
Acknowledgements
We thank R. Sendtner, H. Troll, E. Spirk, and N. Rachor for skillful technical assistance, J. Rieckmann, M. Moradi and P. Lüningschör for discussions, R. Rush from Flinders University, Adelaide, Australia for donating the p75NTR antibody and R. Blum from the Institute of Clinical Neurobiology, University of Wuerzburg for the GFP-actin construct and advice. This work was supported by the European Community′s Health Seventh Framework Programme (FP7) under grant agreement no. 259867 (M.S., P.J.S.), the Hermann-und-Lilly Schilling Stiftung im Stifterverband der Deutschen Industrie (M.S.), a grant by the Deutsche Gesellschaft für Muskelerkrankungen, IBC He 2/2 (A. Hermann, M.S.), The Bavarian Excellence Program ForIPS (M.S.), The DFG SPP 1738 (M.S.), the MeDDrive of the Medical Faculty of the Technische Universität Dresden (A. Hermann), BIOCREA GmbH to A. Hermann, the Helmholtz Virtual Institute program RNA Dysmetabolism in ALS, NOMIS Foundation (A. Hermann) and FTD (VI-510) (A. Hermann), and an unrestricted grant by a family of a deceased ALS patient (A. Hermann).
Author information
Authors and Affiliations
Contributions
R.S., M.S., C.D., D.H., F.M. and M.M. designed the experiments. R.S. developed lentiviruses, performed all experiments to characterize the function of C9ORF72 in cultured motor neurons. C.D. and A. Hansel helped with the initial generation of viral vectors for C9ORF72-HA overexpression and knockdown. D.H. did the LC-MS experiments and D.H., M.F. and M.M. were responsible for the analysis of the LC-MS results. N.F. helped with live cell imaging and analysis. S.J. collected and contributed the lymphoblastoid cell lines. A. Hermann, J.S. and X.L. contributed iPSCs and performed experiments with iPSC-derived motor neurons shown in Supplementary Figure 6. P.J.S. and P.G.I. collected and provided post-mortem tissues from ALS patients. R.S. and M.S. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
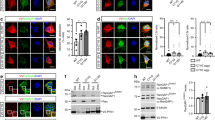
Supplementary Figure 1 Confirmation of C9ORF72 overexpression and knockdown in cultured mouse motor neurons.
A) Diagram of hu-C9ORF72 overexpression vector. (B) Diagram of C9ORF72 shRNA vector with co-expression of GFP under the CMV promoter. (C) Quantification of C9ORF72 RNA expression levels by qPCR in motor neurons infected with and without lentivirus with scrambled shRNA and C9ORF72 shRNA (ANOVA with Bonferroni posthoc test, mean ± s.e.m., F (2, 21)=3.98, p=>0.999, p=<0.001, p=<0.001, n=6 independent experiments). (D) Protein quantification of C9ORF72 protein levels in cultured motor neurons after overexpression or knockdown of C9ORF72. This figure shows one representative blot from n=6 independent experiments. (E) Quantification of the western blots, as shown in D. n=6 independent experiments. F (2, 21)=3.98, p=>0.999, p=<0.001, p=<0.001 ***, P < 0.001; ANOVA with Bonferroni posthoc test, mean ± s.e.m.
Supplementary Figure 2 Altered C9ORF72 expression does not affect motor neuron survival, and overexpression of human C9ORF72 can rescue the axon length defect after knockdown of endogenous mouse C9ORF72.
A) Survival of cultured mouse motor neurons transduced with lentiviral vectors for C9ORF72 overexpression or knockdown, as indicated. Cells were cultured with or without 5ng/ml BDNF, as indicated. Graph shows data from n=4 independent experiments, 100 cells per condition assayed. Kruskal-Wallis statistic= 31.7, p=0.0126, p=>0.999, p=0.0207, p=>0.999, p=0.0374, p=>0.999, p=0.0089. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ANOVA with Kruskal-Wallis test, mean ± s.e.m. (B) Representative image of control motor neurons or motor neurons after lentiviral C9ORF72 overexpression or knockdown at 7d in vitro in the presence of BDNF, stained for tubulin to visualize processes. Bars, 100μm. (C) Human C9ORF72-HA overexpression rescues defective axon elongation caused by knockdown of mouse C9ORF72 after 7d in vitro culture (ANOVA with Bonferroni posthoc test, The central line represents the median, the box limits the interquartile range, and the whiskers the minimum and maximum, F (4, 414) =21.93, p= < 0.001, p=<0.001, p=<0.001, p=>0.999, p=0.935, *** P<0.001, n=4 independent experiments, 100 cells per condition assayed). NS: not significant.
Supplementary Figure 3 Interactome of C9ORF72 in NSC34 cells.
(A) Western blot analysis of HEK293 and NSC-34 cells overexpressing huC9ORF72-HA. (B) Western blot analysis of immunoprecipitates of huC9ORF72-HA protein. Transduced huC9ORF72 was precipitated with antibodies against HA, resulting blots were stained with antibodies against huC9ORF72. (C) Confirmative blot of samples used for LC-MS analysis. Samples were pulled down with HA antibody and the blots were probed with C9ORF72 antibody. 1% of immunoprecipitation sample and 10% of input was used for the analysis.
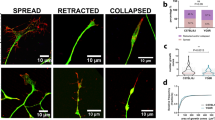
Supplementary Figure 4 Interaction of C9ORF72 and cofilin in motor neurons.
(A) HA pulldown of C9ORF72-HA protein from motor neurons cultured for 7d in vitro and immunoblotting for C9ORF72 and cofilin. (B) Immunoprecipitation of endogenous C9ORF72 from cultured motor neurons. The resulting immunoblot exposed to cofilin antibodies confirms the interaction of C9ORF72 with cofilin. (C) Motor neurons after 7d in vitro stained for C9ORF72-HA, cofilin and phalloidin. Bars 100μm. (D) Colocalization of C9ORF72-HA with cofilin in axonal growth cones of motor neurons grown for 5d in vitro. Bars 10μm.
Supplementary Figure 5 Construction and efficacy of viral constructs for C9ORF72 knockdown and coexpression of GFP-actin.
(A) Representative scheme of the C9ORF72 shRNA vector with co-expression of GFP-actin under the CMV promoter. (B) Motor neurons transduced with C9ORF72 shRNA or scrambled shRNA lentiviruses were detected at 7d in vitro by co expressed GFP-actin. GFP-actin co-localizes with actin and tubulin in axonal processes. Bars 100μm. (C) Western blot analysis of cultured motor neurons at 7d in vitro transduced with shRNA for scrambled and C9ORF72 with coexpression of GFP and GFP-actin. The blot also shows the effect of knockdown on C9ORF72 protein levels when extracts from cells transduced with shRNA viruses are compared with cells carrying control scrambled constructs.
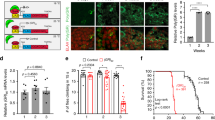
Supplementary Figure 6 Characterization of the C9ORF72 patient-specific iPSC cell lines.
(A) C9ORF72 iPSC colonies express characteristic pluripotency markers as shown by an immunofluorescence staining of Oct4, TRA-1-60 and TRA-1-81. (B) C9ORF72 iPSCs (#1=patient1; #2=patient2) are successfully silenced upon reprogramming. FIB, fibroblasts (C) C9ORF72 cells successfully differentiate into cells from all there germ layers as shown by the expression of the germ layer markers Catenin (Endoderm), alpha-SMA/FN (Mesoderm; FN= fibronectin) and TUJ1 (Ectoderm). Scale bars 50µm.
Supplementary Figure 7 Immunocytochemical characterization of iPSC neurons.
(A) iPSCs differentiated into motor neuron include phenotypic markers such as Islet-1, HB9, Tuj1, MAP2 both in control and C9-ALS derived neurons. Bars 100μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 1717 kb)
Supplementary Table 1
Protein table: List of identified C9ORF72 interactors (XLSX 330 kb)
Supplementary Table 2
GO term analysis of identified C9ORF72 interactors (XLSX 11 kb)
41593_2016_BFnn4407_MOESM59_ESM.avi
Live cell imaging of the axonal growth cone in a human control iPS cell derived motoneuron transduced with GFP-actin sh scrambled (AVI 675 kb)
41593_2016_BFnn4407_MOESM60_ESM.avi
Live cell imaging of the axonal growth cone in a human C9-ALS iPS cell derived motoneuron transduced with GFP-actin sh scrambled (AVI 766 kb)
Rights and permissions
About this article
Cite this article
Sivadasan, R., Hornburg, D., Drepper, C. et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci 19, 1610–1618 (2016). https://doi.org/10.1038/nn.4407
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4407
This article is cited by
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
Roadmap for C9ORF72 in Frontotemporal Dementia and Amyotrophic Lateral Sclerosis: Report on the C9ORF72 FTD/ALS Summit
Neurology and Therapy (2023)
-
Therapy development for spinal muscular atrophy: perspectives for muscular dystrophies and neurodegenerative disorders
Neurological Research and Practice (2022)
-
Autophagy Dysfunction in ALS: from Transport to Protein Degradation
Journal of Molecular Neuroscience (2022)
-
An interaction between synapsin and C9orf72 regulates excitatory synapses and is impaired in ALS/FTD
Acta Neuropathologica (2022)